4. Visualization¶
This page introduces the commands in CustardPy to plot the 3D visualization.
These commands use the output data generated by custardpy_juicer or custardpy_process_hic.
Here we use the example data used in Step-by-Step Workflow of Hi-C Analysis.
4.1. plotHiCMatrix¶
plotHiCMatrix visualizes a contact map as a simple square heatmap. The input data is a dense matrix output from makeMatrix_intra.sh.
The contact level is normalized by the total number of mapped reads for the chromosome.
plotHiCMatrix <matrix> <output name> <start> <end> <title in figure>
Example:
chr=chr20
start=8000000
end=16000000
resolution=25000
norm=SCALE
matrix=CustardPyResults_Hi-C/Juicer_hg38/Control/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz
plotHiCMatrix \
$matrix \
ContactMap.Control.$chr.$start-$end.pdf \
$start $end Control
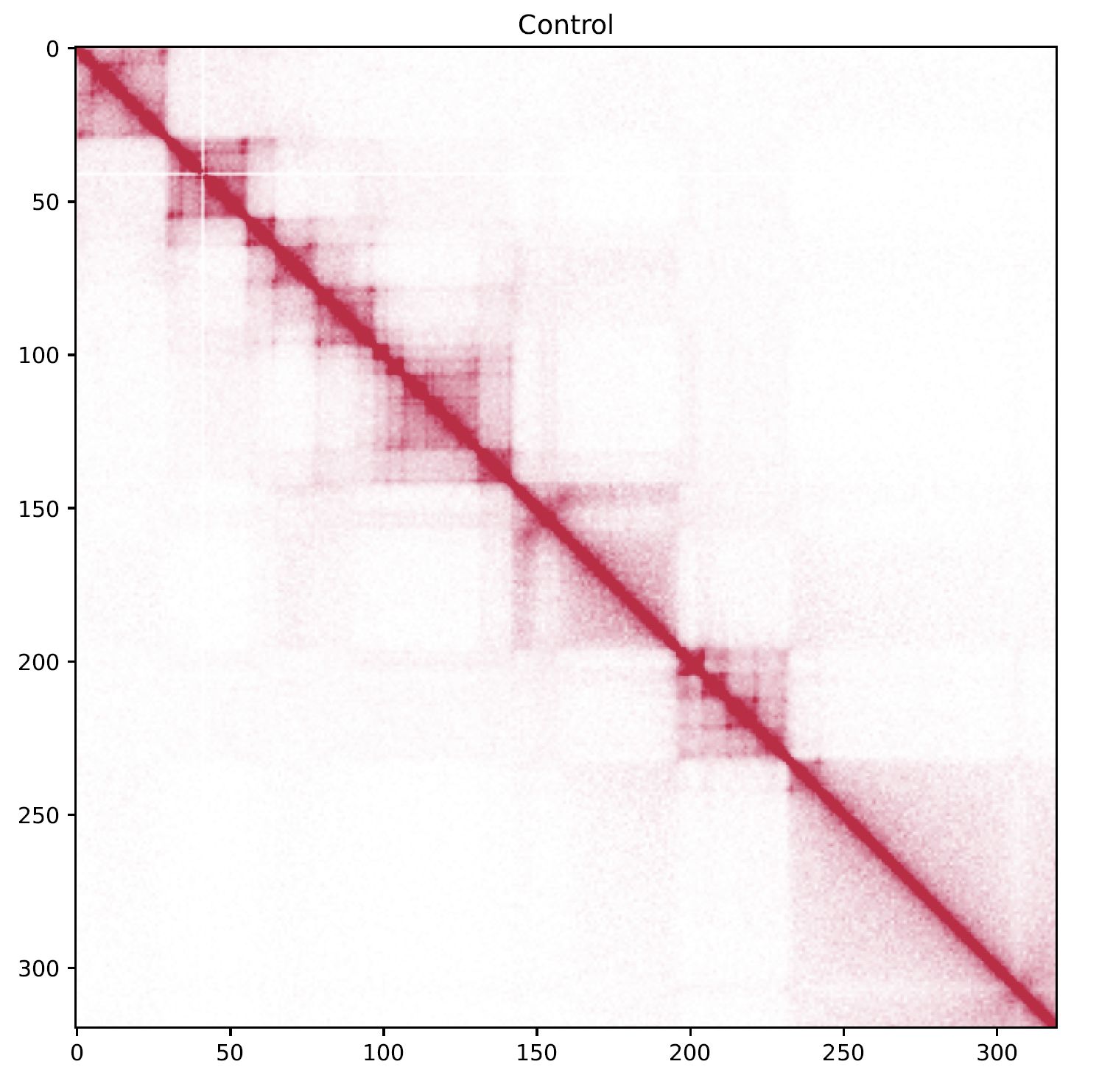
Fig. 4.1 plotHiCMatrix¶
4.2. drawSquareMulti¶
drawSquareMulti visualizes the contact map of multiple Hi-C samples as triangle heatmaps.
The input data is a dense matrix output from makeMatrix_intra.sh.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
# linear scale
drawSquareMulti \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o SquareMulti.$chr \
-c $chr --start $start --end $end --type $norm -r $resolution
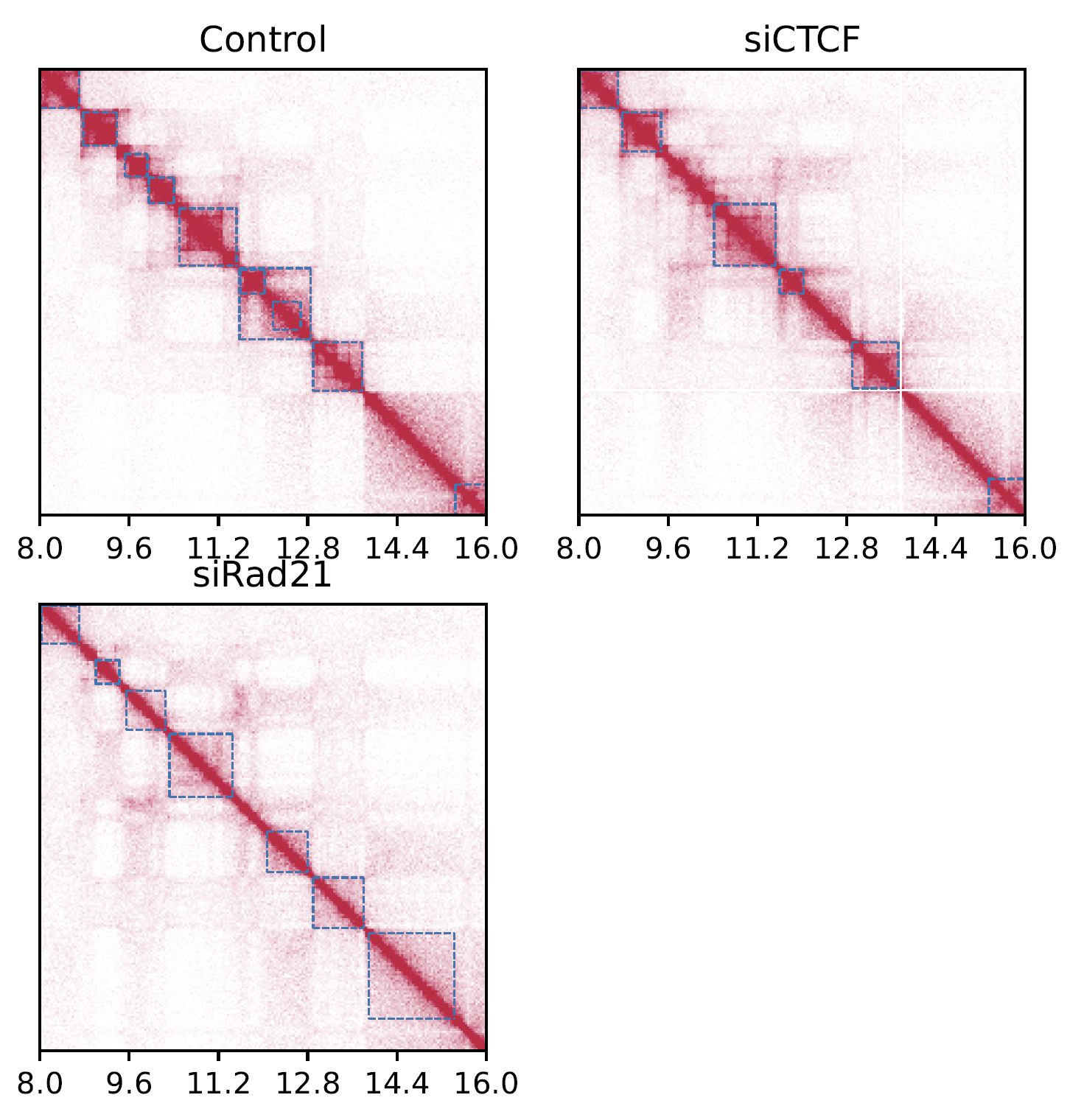
Fig. 4.2 SquareMulti¶
The dashed squares indicate TADs.
Add --log option to visualize a log-scale heatmap.
# log scale
drawSquareMulti \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o SquareMulti.$chr \
-c $chr --start $start --end $end \
--type $norm -r $resolution --log
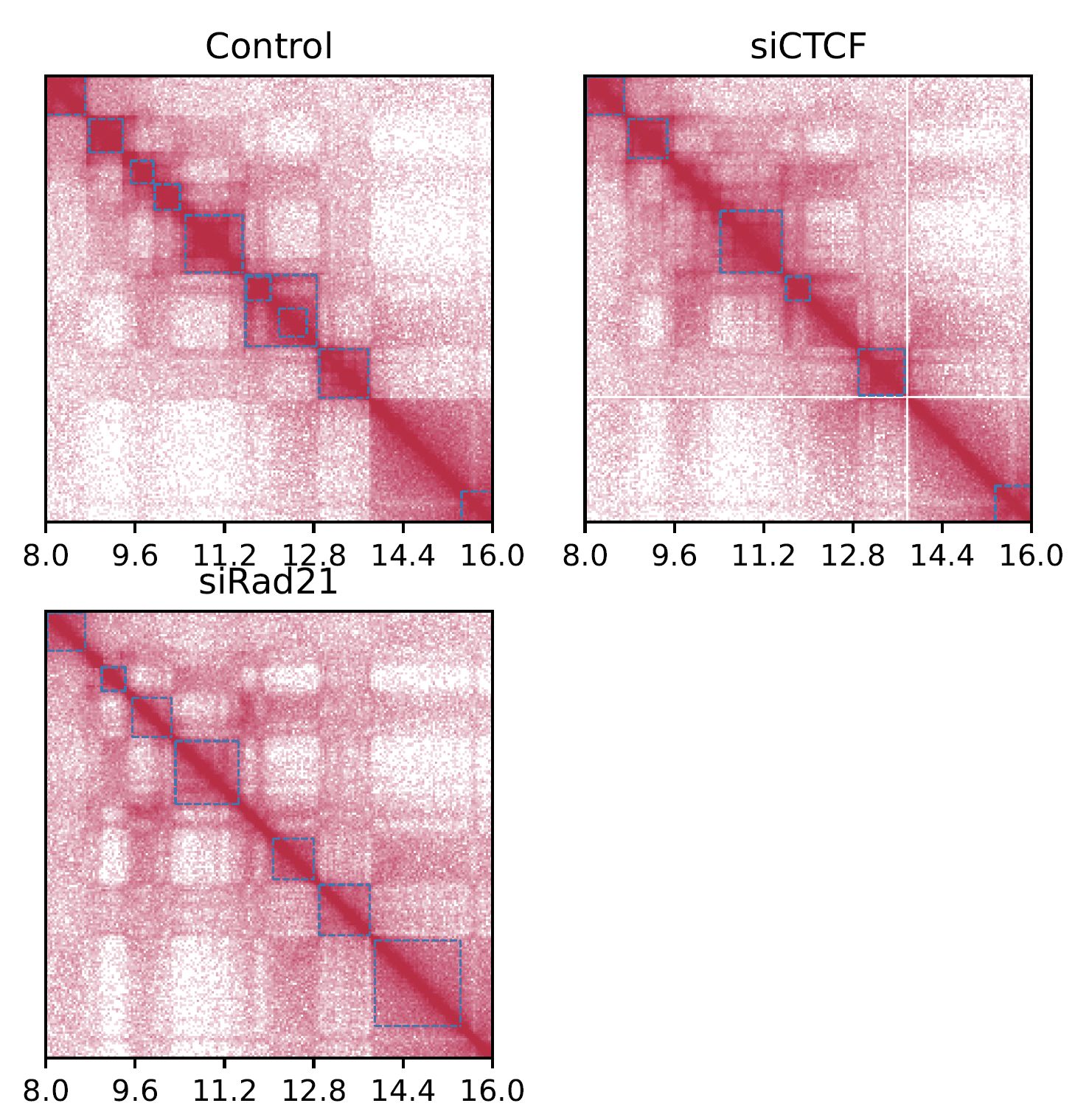
Fig. 4.3 SquareMulti (log scale)¶
4.3. drawSquareRatioMulti¶
drawSquareRatioMulti visualizes a relative contact frequency (log scale) of 2nd to the last samples against the first sample.
The input data is a dense matrix output from makeMatrix_intra.sh.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
drawSquareRatioMulti \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o SquareRatioMulti.$chr \
-c $chr --start $start --end $end --type $norm -r $resolution
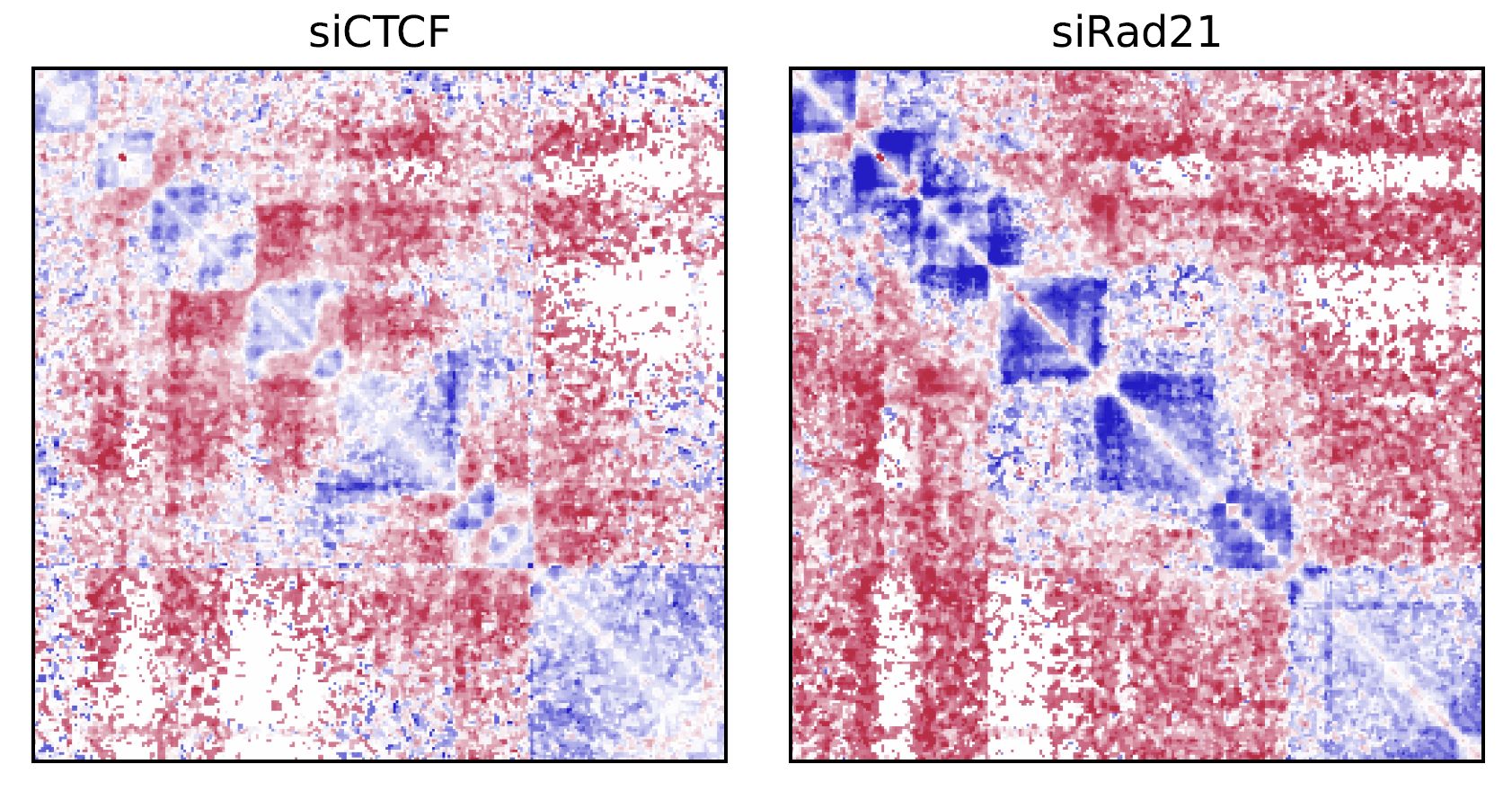
Fig. 4.4 drawSquareRatioMulti¶
This figure shows the relative contact frequency of 2nd (siCTCF) and 3rd (siRad21) against 1st (Control).
4.4. drawSquarePair¶
drawSquarePair shows two samples in a single square heatmap.
The first and second samples are visualzed in the upper and bottom triagles, respectively.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
drawSquarePair \
$Resdir/Control/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:Control \
$Resdir/siRad21/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:siRad21 \
-o SquarePair.$chr --start $start --end $end -r $resolution
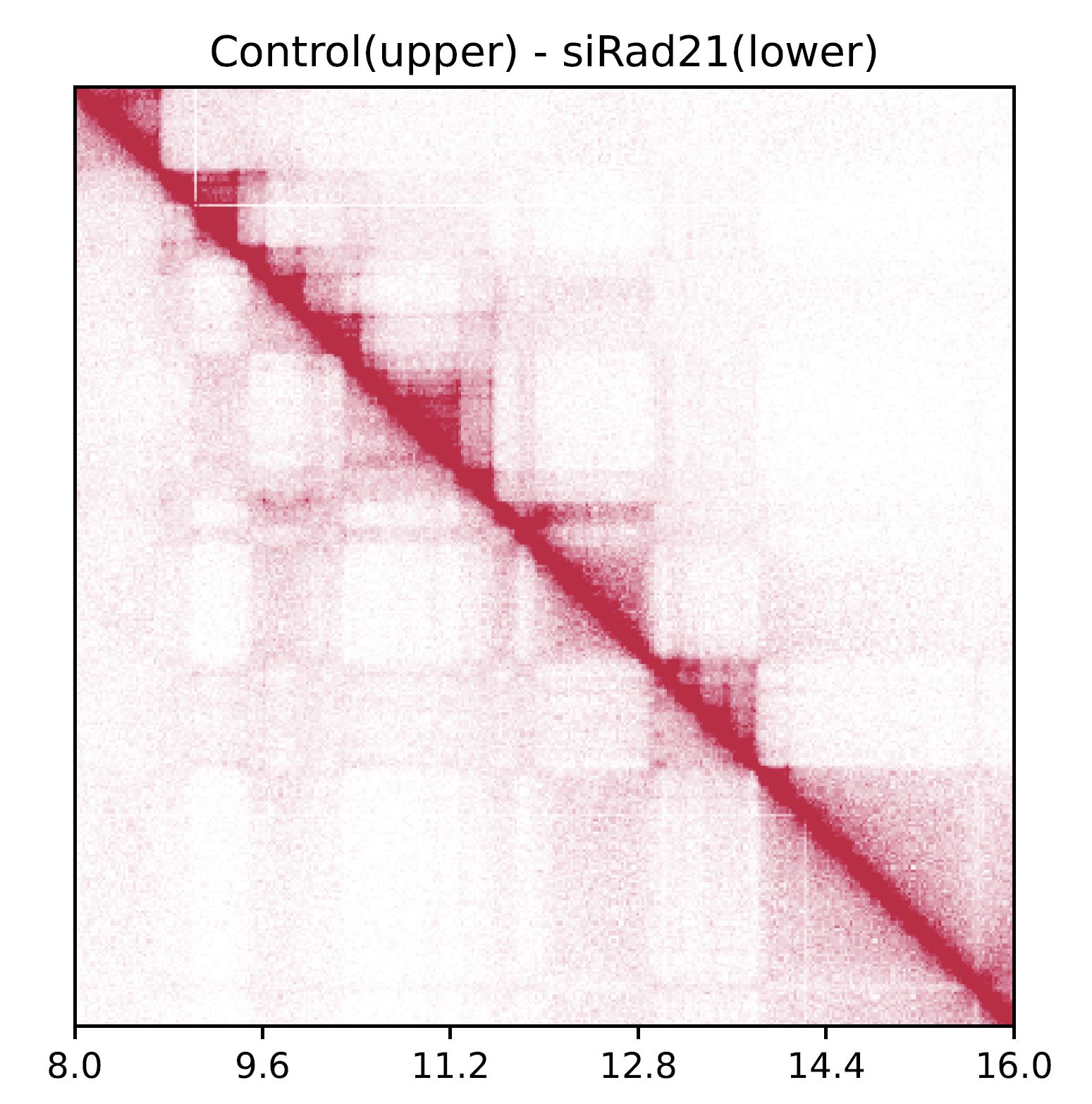
Fig. 4.5 drawSquarePair¶
The command visualizes Control and siRad21 in the upper and bottom triagles, respectively.
4.5. drawSquareRatioPair¶
Similar to drawSquarePair, drawSquareRatioPair shows the relative contact map of two sample pairs in a single square heatmap.
This command visualize the log-scale frequency of sample2/sample1 and sample4/sample3 in the upper and bottom triagles, respectively.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
drawSquareRatioPair \
$Resdir/Control/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:Control \
$Resdir/siRad21/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:siRad21 \
$Resdir/Control/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:Control \
$Resdir/siCTCF/Matrix/intrachromosomal/$resolution/observed.$norm.$chr.matrix.gz:siCTCF \
-o drawSquareRatioPair.$chr --start $start --end $end -r $resolution
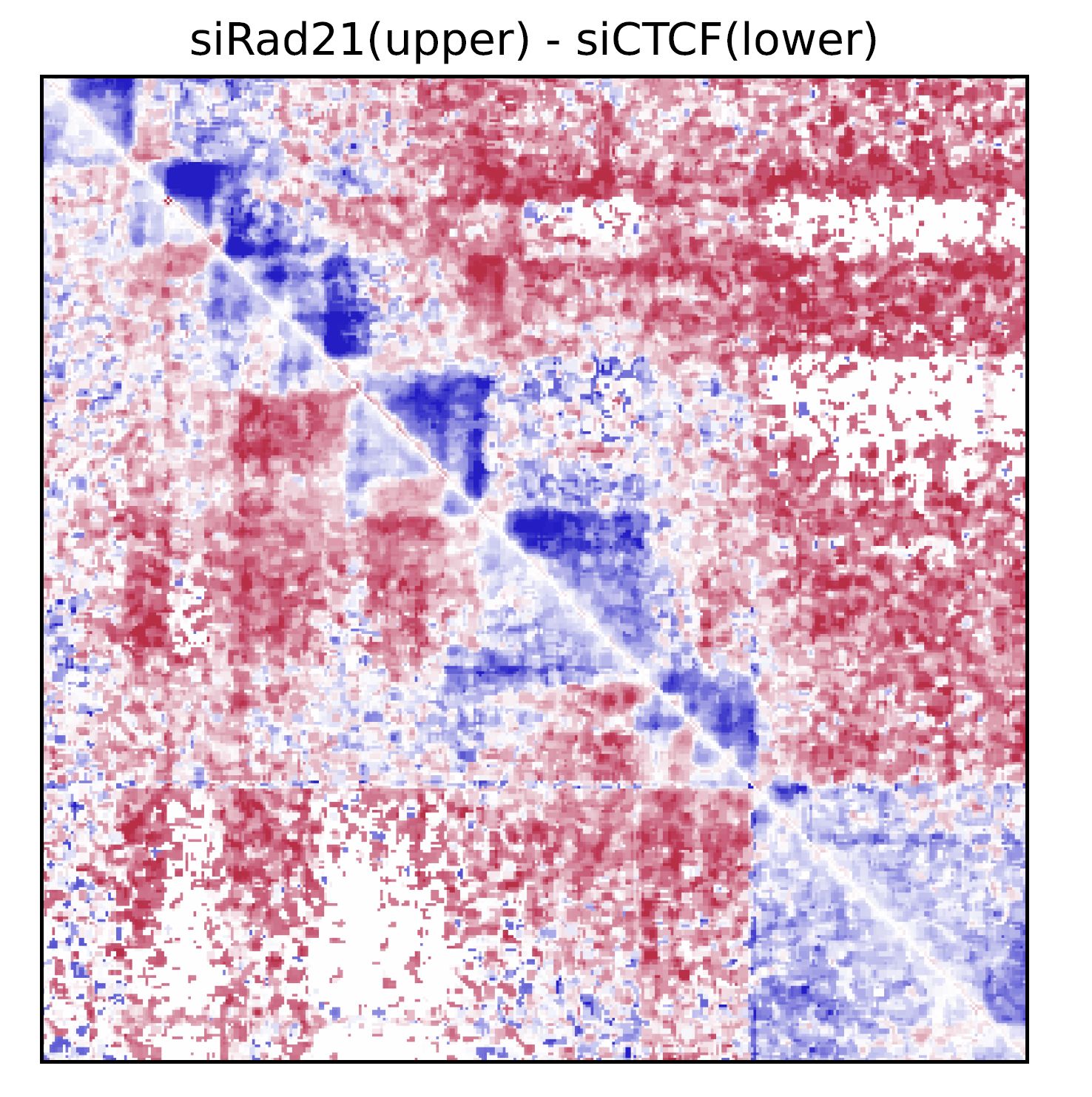
Fig. 4.6 drawSquareRatioPair¶
In this case, siRad21/Control and siCTCF/Control are visualized in the upper and bottom triagles, respectively.
4.6. drawTriangleMulti¶
drawTriangleMulti visualizes the contact map of multiple Hi-C samples as triangle heatmaps.
The input data is a dense matrix output from makeMatrix_intra.sh.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
drawTriangleMulti \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o TriangleMulti.$chr \
-c $chr --start $start --end $end --type $norm -d 5000000 -r $resolution
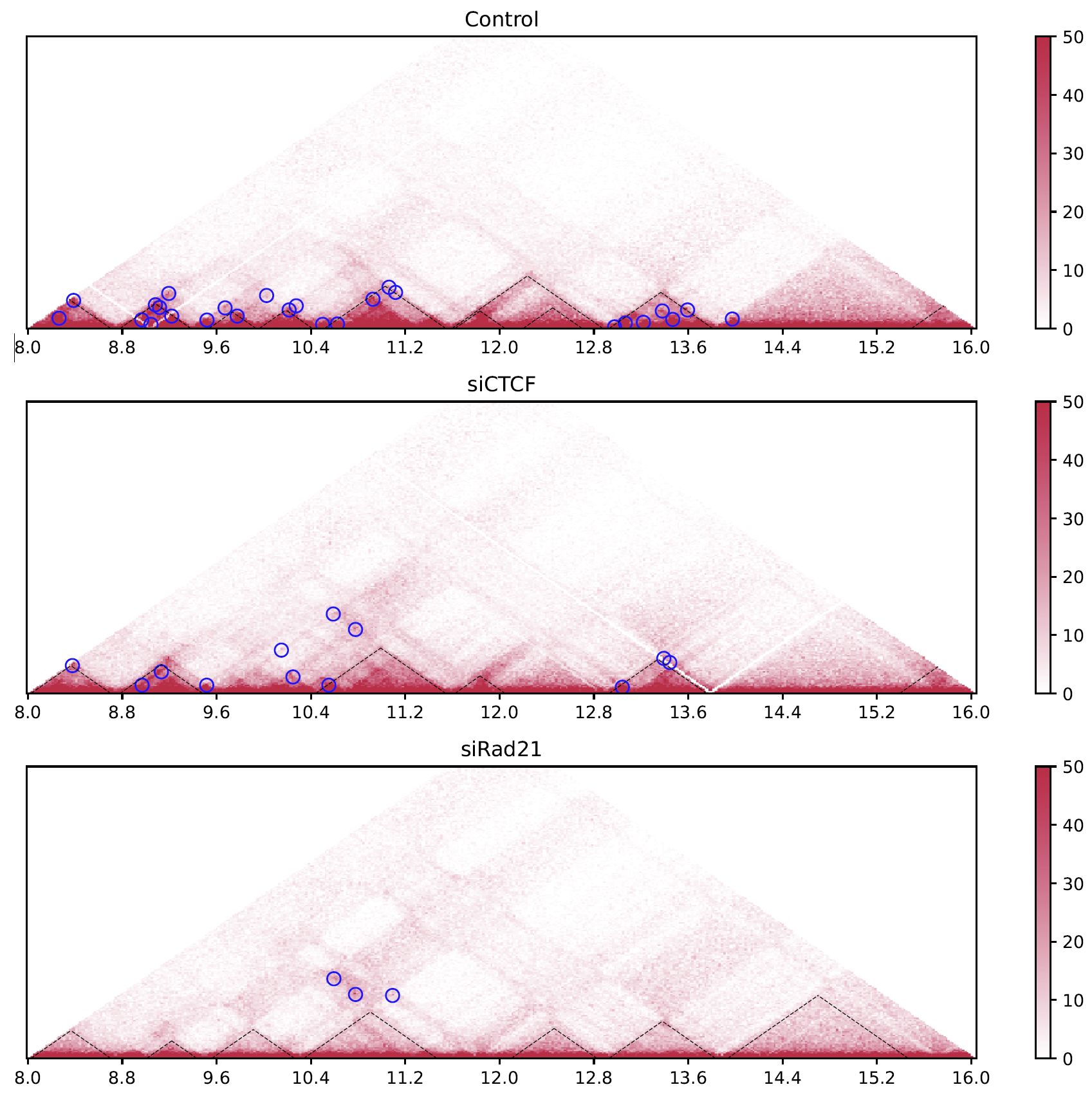
Fig. 4.7 drawTriangleMulti¶
The black dashed lines and blue circles indicate TADs and loops, respectively.
4.7. drawTrianglePair¶
drawTrianglePair visualizes a contact map of the first and second sample in upper and lower triangles, respectively.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
drawTrianglePair \
$Resdir/Control:Control \
$Resdir/siRad21:siRad21 \
-o TrianglePair.$chr \
-c $chr --start $start --end $end --type $norm -d 5000000 -r $resolution
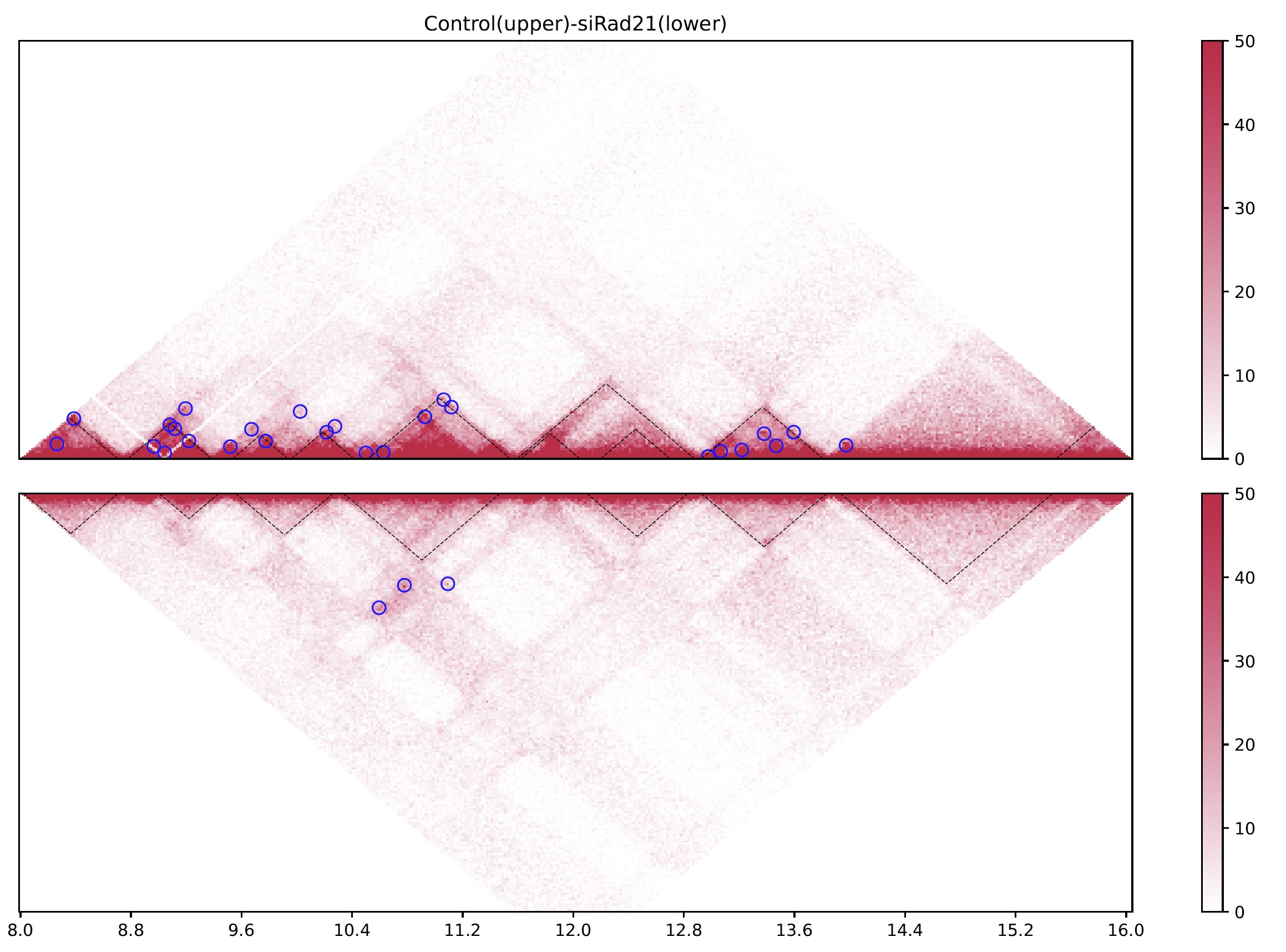
Fig. 4.8 drawTrianglePair¶
The black dashed lines and blue circles indicate TADs and loops, respectively.
4.8. plotHiCfeature¶
plotHiCfeature can visualize various features values of chromatin folding for multiple samples.
plotHiCfeature [-h] [-o OUTPUT] [-c CHR] [--type TYPE]
[--distance DISTANCE] [-r RESOLUTION] [-s START]
[-e END] [--multi] [--multidiff] [--compartment] [--di]
[--drf] [--drf_right] [--drf_left]
[--triangle_ratio_multi] [-d VIZDISTANCEMAX] [--v4c]
[--vmax VMAX] [--vmin VMIN] [--vmax_ratio VMAX_RATIO]
[--vmin_ratio VMIN_RATIO] [--anchor ANCHOR]
[--xsize XSIZE]
[input [input ...]]
positional arguments:
input <Input directory>:<label>
optional arguments:
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Output prefix
-c CHR, --chr CHR chromosome
--type TYPE normalize type (default: SCALE)
--distance DISTANCE distance for DI (default: 500000)
-r RESOLUTION, --resolution RESOLUTION
resolution (default: 25000)
-s START, --start START
start bp (default: 0)
-e END, --end END end bp (default: 1000000)
--multi plot MultiInsulation Score
--multidiff plot differential MultiInsulation Score
--compartment plot Compartment (eigen)
--di plot Directionaly Index
--drf plot Directional Relative Frequency
--drf_right (with --drf) plot DirectionalRelativeFreq (Right)
--drf_left (with --drf) plot DirectionalRelativeFreq (Left)
--triangle_ratio_multi
plot Triangle ratio multi
-d VIZDISTANCEMAX, --vizdistancemax VIZDISTANCEMAX
max distance in heatmap
--v4c plot virtual 4C from Hi-C data
--vmax VMAX max value of color bar (default: 50)
--vmin VMIN min value of color bar (default: 0)
--vmax_ratio VMAX_RATIO
max value of color bar for logratio (default: 1)
--vmin_ratio VMIN_RATIO
min value of color bar for logratio (default: -1)
--anchor ANCHOR (for --v4c) anchor point
--xsize XSIZE xsize for figure (default: max(length/2M, 10))
Inputshould be “<sample directory>:<label>”.<sample directory>is the output directory bycustardpy_juicer.<label>is the label used in the figure.
In default,
plotHiCfeatureuses a 25-kbp bin matrix. Supply-roption to change the resolution.typeis the normalization type defined by Juicer (SCALE/KR/VC_SQRT/NONE).
4.8.1. Insulation score¶
By default, plotHiCfeature outputs a single insulation score (500 kbp distance).
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--type $norm -d 5000000 \
-o IS.$chr.$start-$end
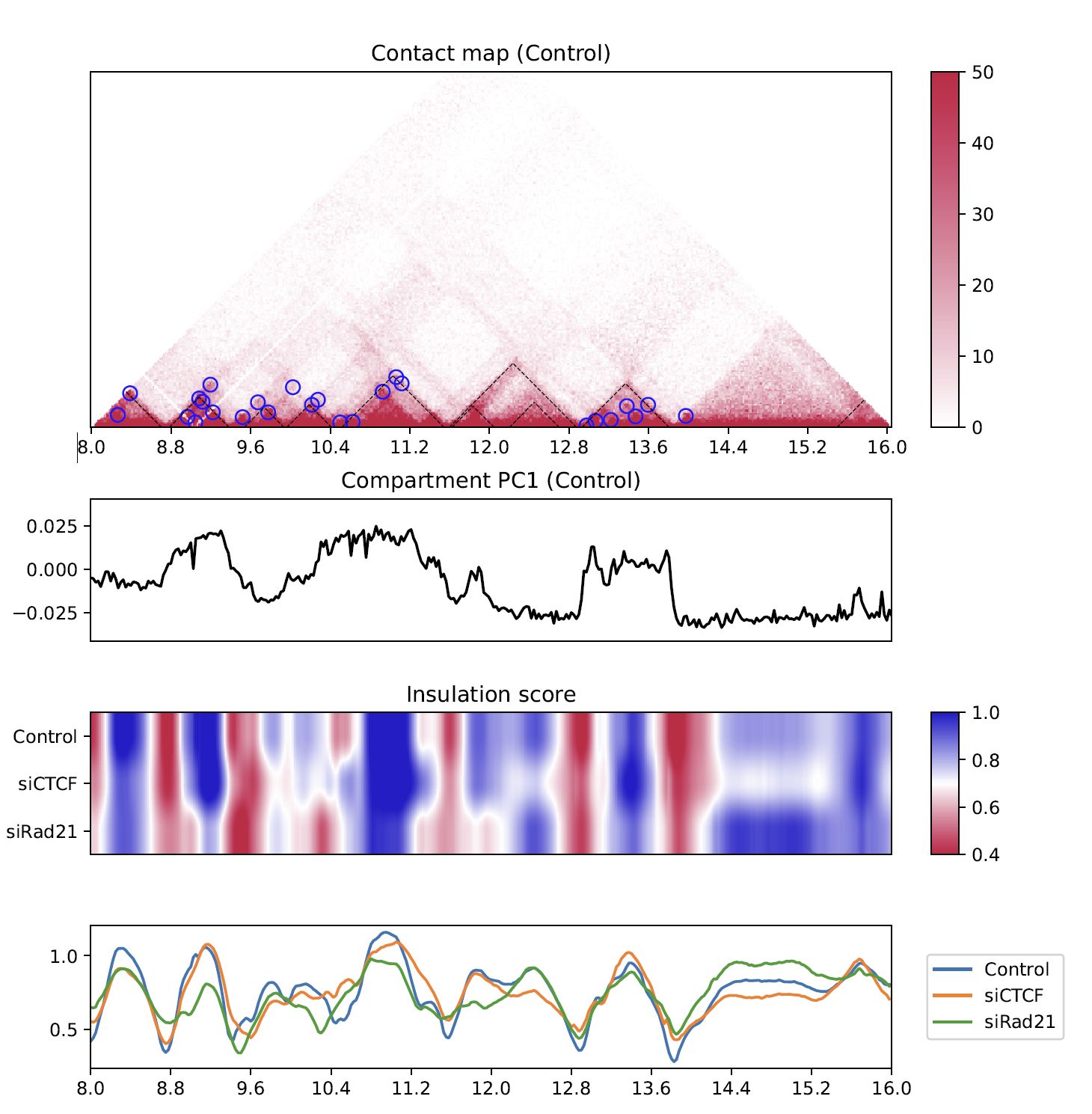
Fig. 4.9 Insulation score¶
plotHiCfeature always draws compartment PC1 (line plot in the second row) to roughly estimate compartments A and B.
The third and fourth rows are the heatmap and line plot for the feature value specified (insulation score in this case).
4.8.2. Multi-insulation score¶
plotHiCfeature can also calculate a “multi-scale insulation score” [Crane et al., Nature, 2015] by supplying --multi option.
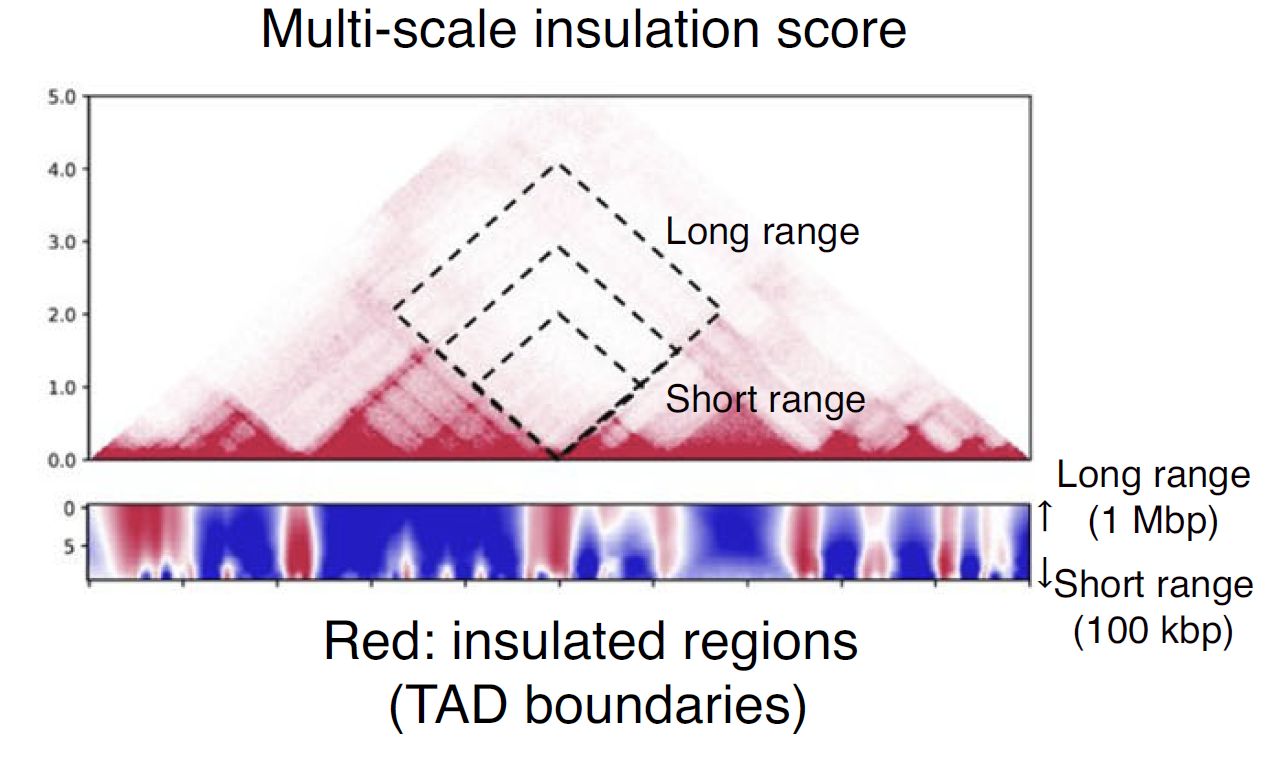
Fig. 4.10 Schematic illustration of multi-insulation score¶
Red regions in the heatmap indicate the insulated regions (TAD boundary-like). The lower and upper sides of the heatmap are 100 kbp to 1 Mbp distances, respectively.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--multi --type $norm -d 5000000 \
-o MultiIS.$chr.$start-$end
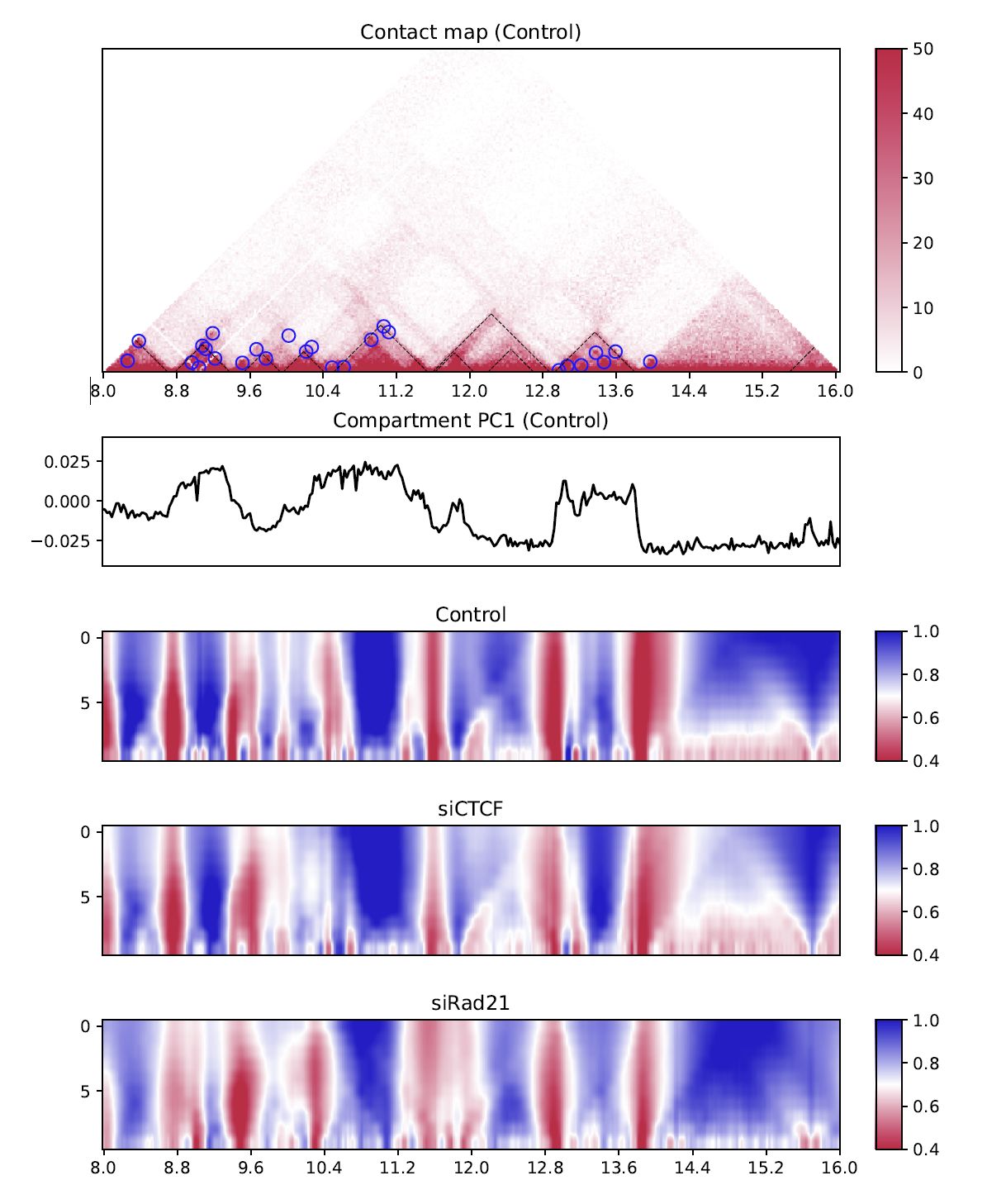
Fig. 4.11 Multi-insulation score¶
4.8.3. Differential multi-insulation score¶
To directory investigate the difference of multi-insulation score, we provide differential multi-insulation score by --multidiff option.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--multidiff --type $norm -d 5000000 \
-o MultiISdiff.$chr.$start-$end
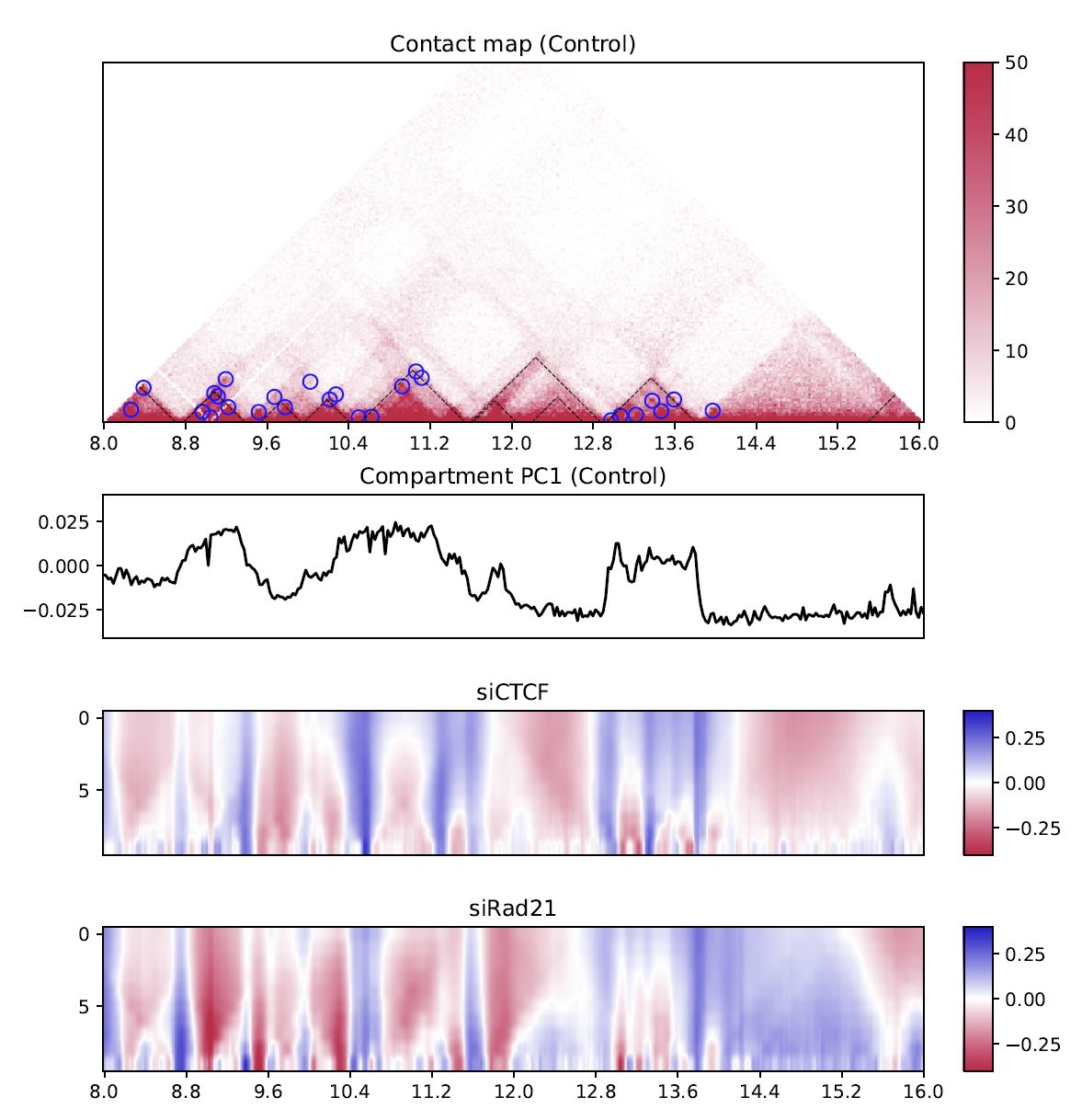
Fig. 4.12 Differential multi-insulation score¶
The heatmap shows the difference against the first sample (siCTCF - Control and siRad21 - Control in this case).
4.8.4. Compartment PC1¶
While the line plot in the second row shows the PC1 value of the first sample,
plotHiCfeature --compartment visualizes the PC1 values for multiple samples.
This plot can be used to explore compartment switching.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--compartment --type $norm -d 5000000 \
-o Compartment.$chr.$start-$end
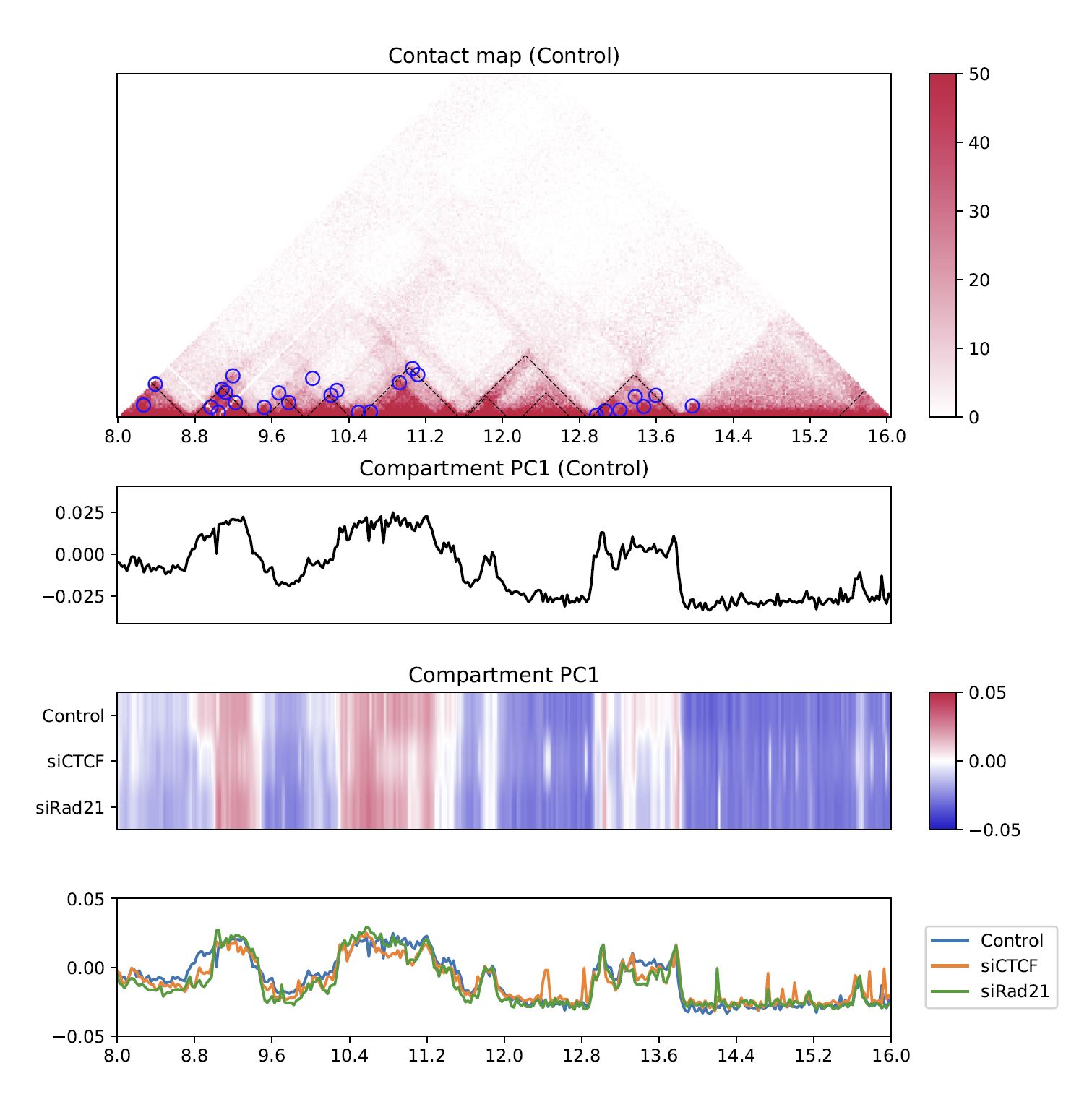
Fig. 4.13 Compartment PC1¶
4.8.5. Directionality index¶
The directionality index identifies TAD boundaries by capturing the bias in contact frequency up- and downstream of a TAD [Dixon et al., Nature, 2012]. The “left side” and “right side” of a TAD are likely to have positve and negative values, respectively.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--di --type $norm -d 5000000 \
-o DI.$chr.$start-$end
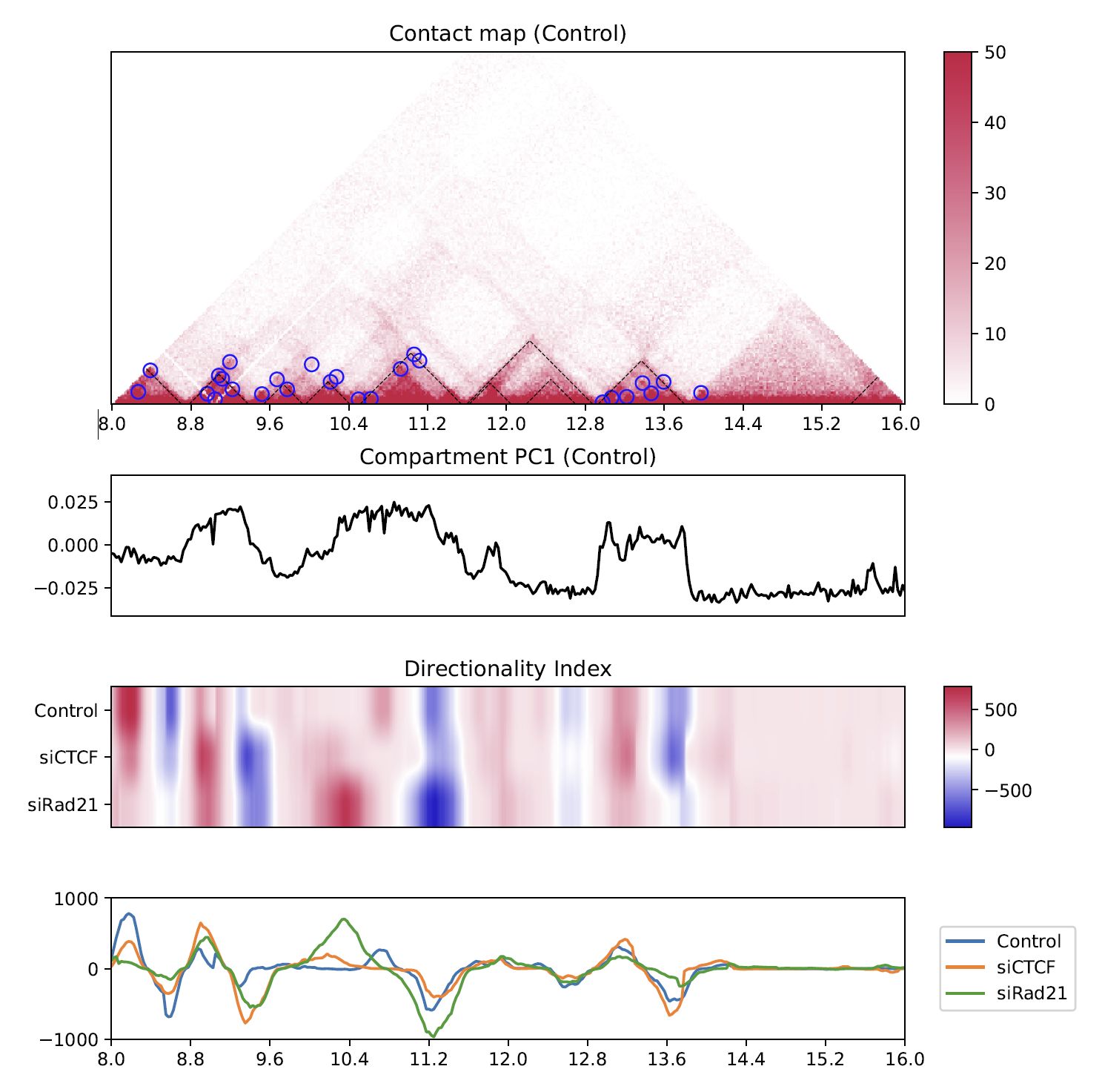
Fig. 4.14 Directionality index¶
4.8.6. Directional relative frequency¶
Directional relative frequency (DRF) is a score that our group previously proposed [Nakato et al, Nature Communications, 2023]. This score estimates the inconsistency of relative contact frequence (log scale) between the left side (3′) and right side (5′).
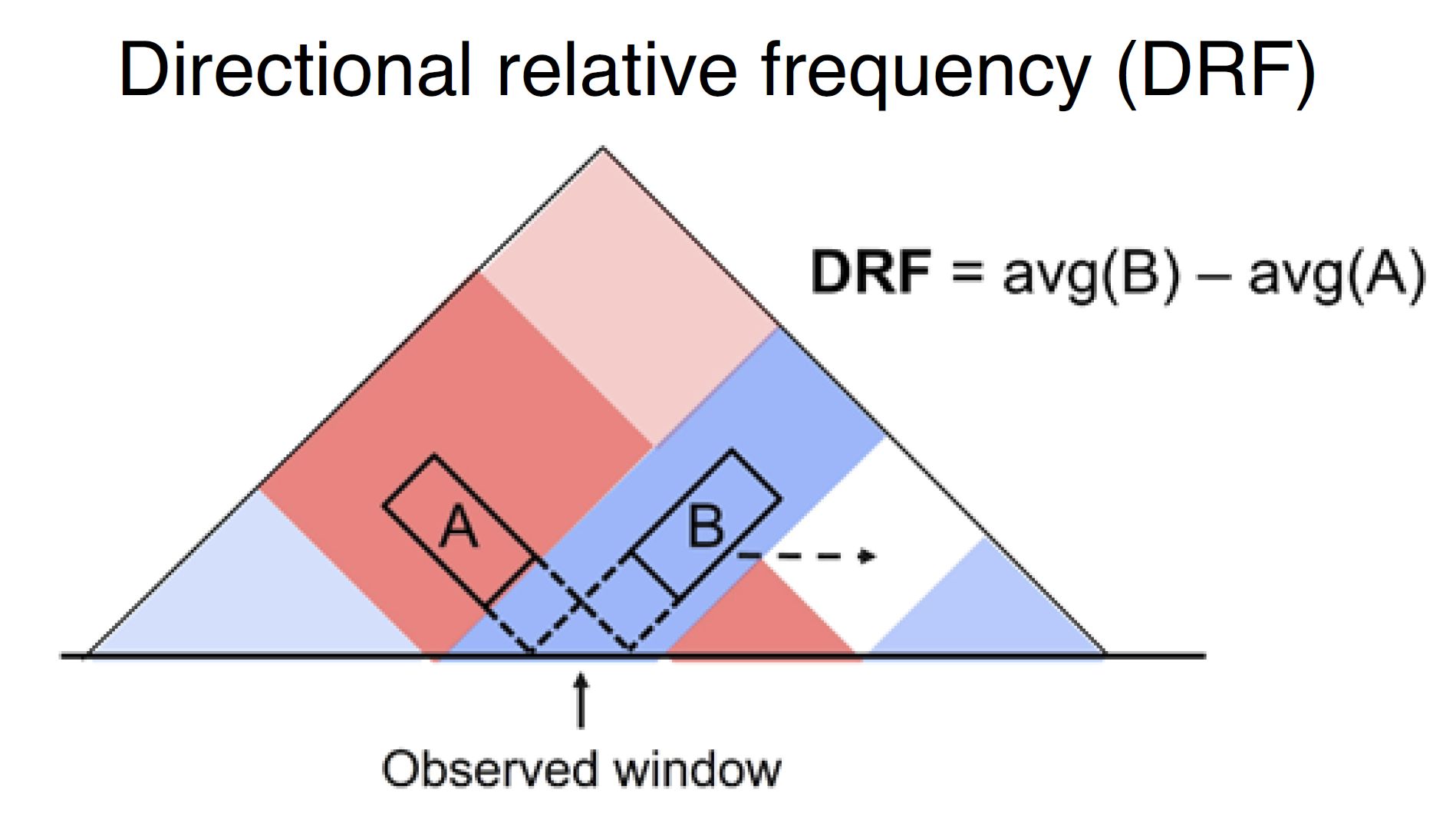
Fig. 4.15 Schematic illustration of directional relative frequency¶
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-c $chr --start $start --end $end -r $resolution \
--drf --type $norm -d 5000000 \
-o DRF.$chr.$start-$end
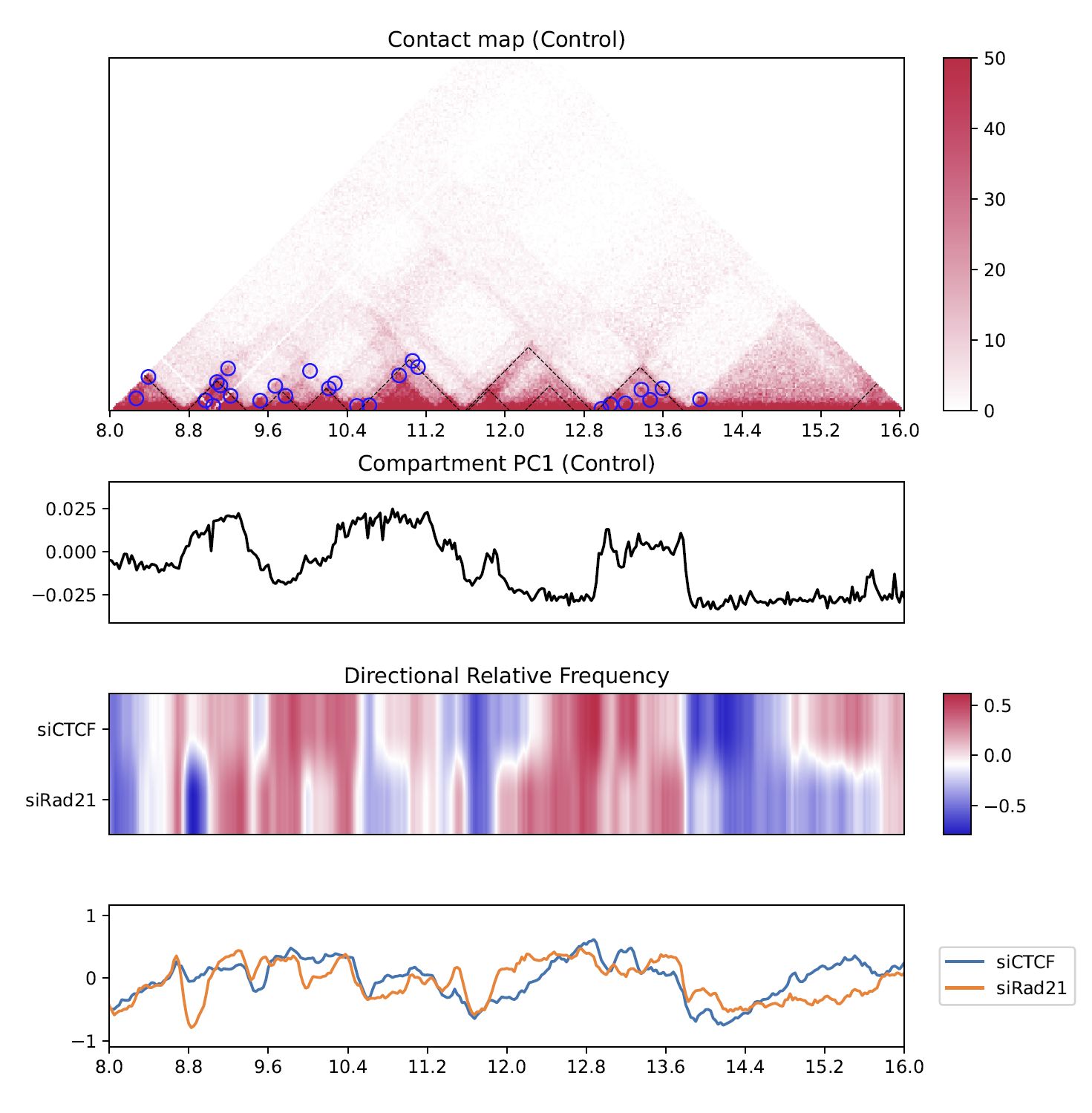
Fig. 4.16 Directional relative frequency¶
4.8.7. drawTriangleRatioMulti¶
drawTriangleRatioMulti visualizes a relative contact frequency (log scale) of 2nd to the last samples against the first sample. Directional relative frequency is also shown.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o TriangleRatioMulti.$chr \
-c $chr --start $start --end $end -r $resolution \
--triangle_ratio_multi --type $norm -d 5000000
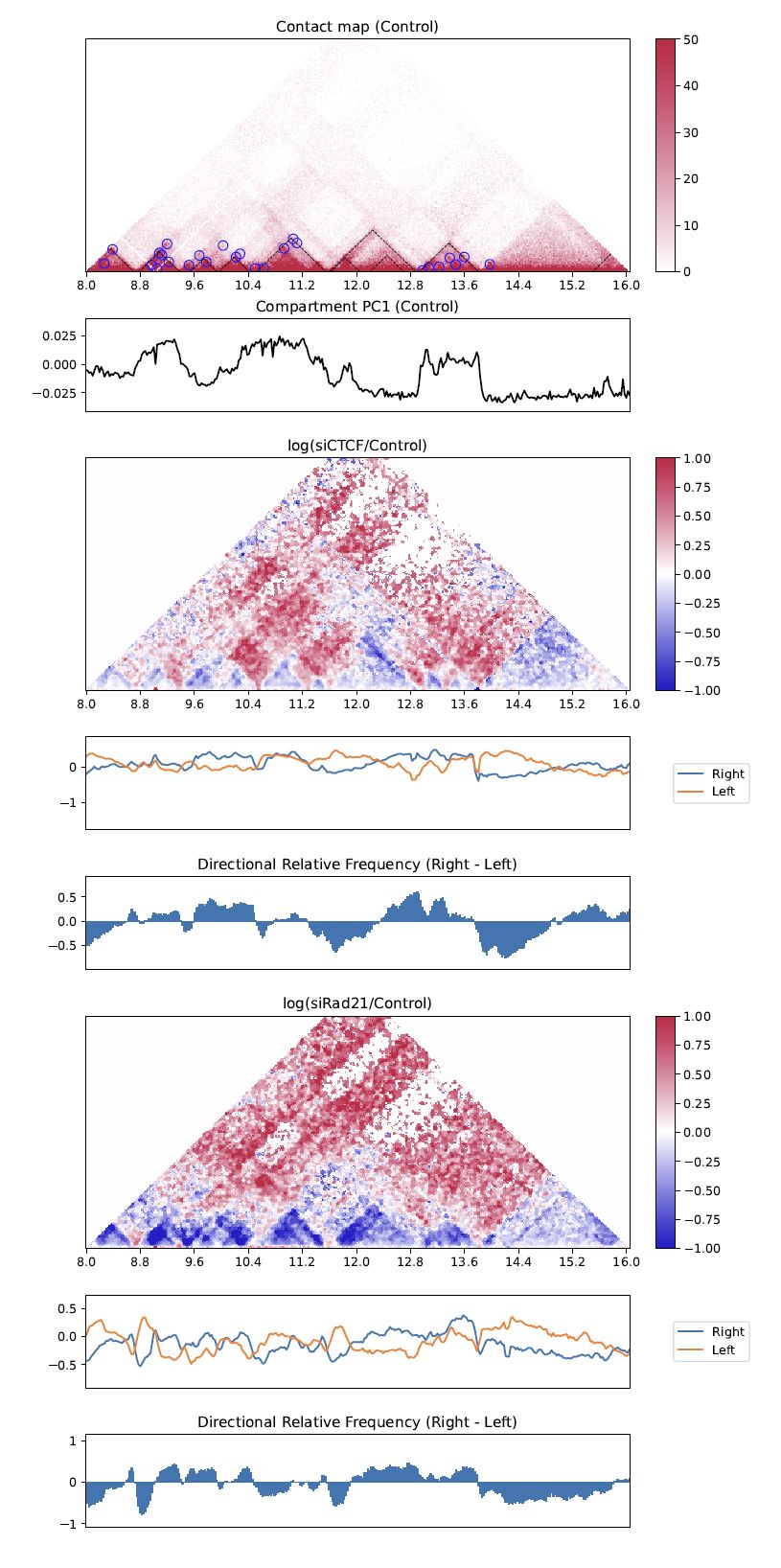
Fig. 4.17 TriangleRatioMulti¶
“Right” and “left” shown as blue and orange line plots in the second row indicate the “B” and “A” in Fig. 4.15.
4.8.8. Virtual 4C¶
Virtual 4C visualizes a 4C-like plot (interation from the anchor site) using Hi-C data.
Use --anchor option to specify the anchor site.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotHiCfeature \
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o virtual4C.$chr \
-c $chr --start $start --end $end -r $resolution \
--v4c --anchor 10400000 --vmax 100 --type $norm
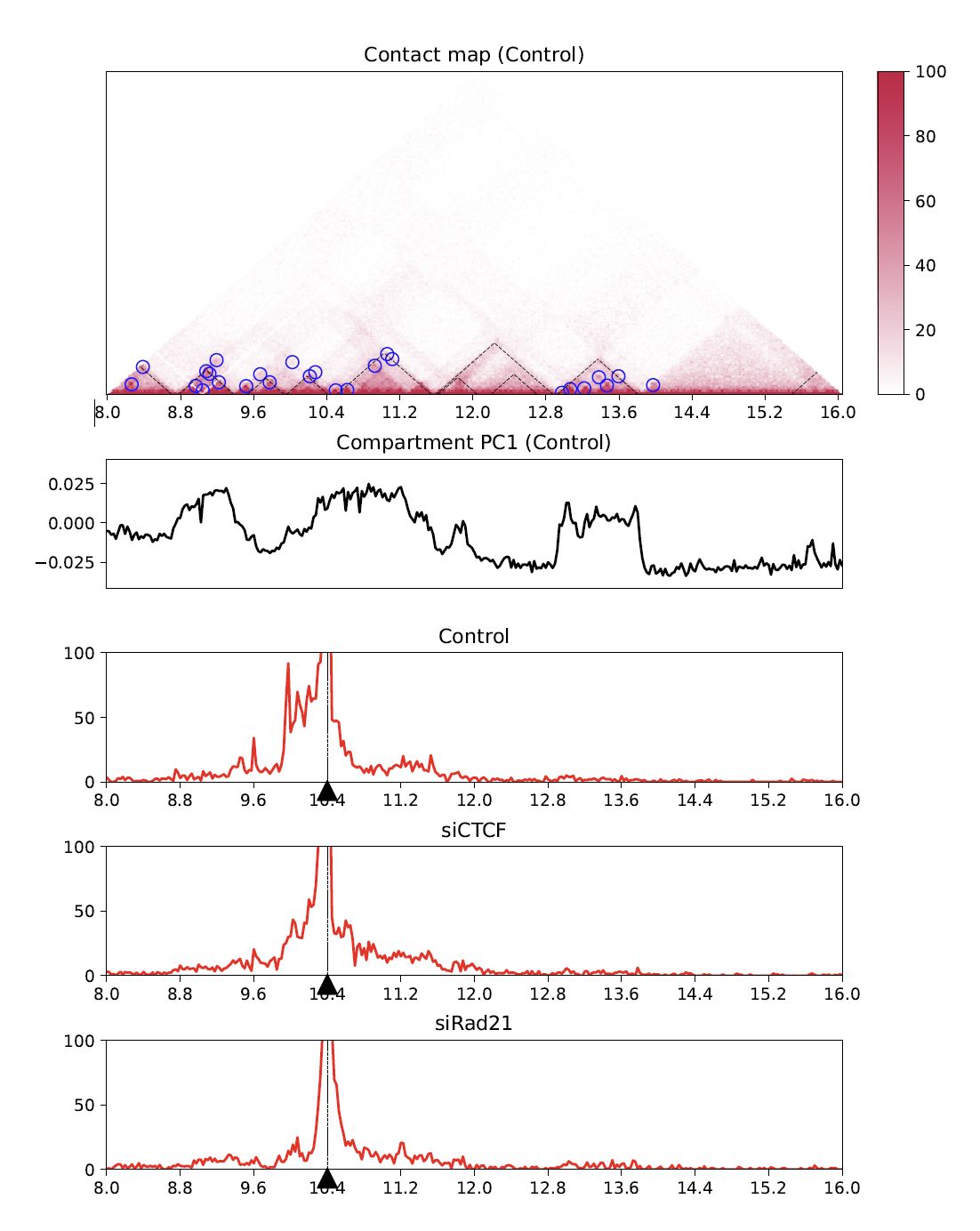
Fig. 4.18 Virtual 4C¶
4.9. plotCompartmentGenome¶
Plot a PC1 value of multiple samples for the whole genome.
Resdir=CustardPyResults_Hi-C/Juicer_hg38
norm=SCALE
resolution=25000
chr=chr20
start=8000000
end=16000000
plotCompartmentGenome
$Resdir/Control:Control \
$Resdir/siCTCF:siCTCF \
$Resdir/siRad21:siRad21 \
-o CompartmentGenome -r 25000 --type VC_SQRT
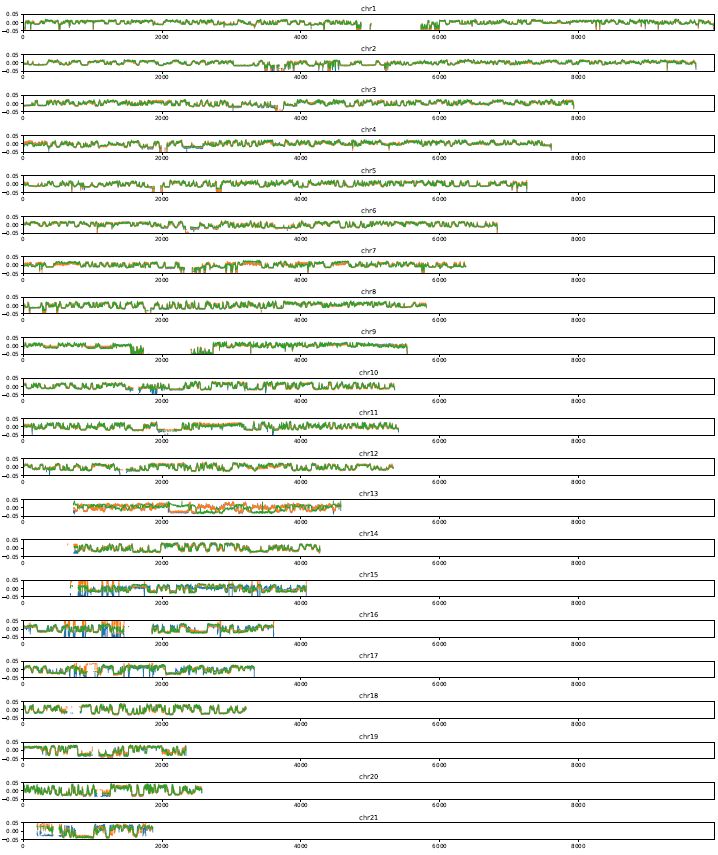
Fig. 4.19 plotCompartmentGenome¶
4.10. plotInsulationScore¶
Plot a line graph of insulation score. The input data is a dense matrix output from makeMatrix_intra.sh.
plotInsulationScore [-h] [--num4norm NUM4NORM] [--distance DISTANCE]
[--sizex SIZEX] [--sizey SIZEY]
matrix output resolution
Example:
plotInsulationScore WT/intrachromosomal/25000/observed.KR.chr7.matrix.gz InsulationScore_WT.chr7.png 25000
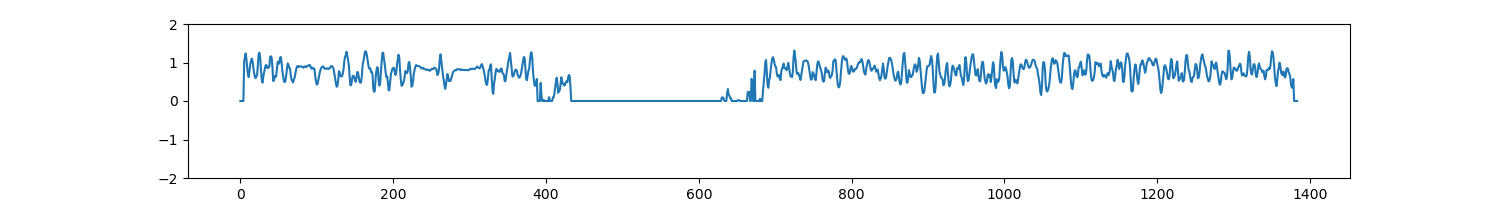
Fig. 4.20 InsulationScore¶
4.11. plotMultiScaleInsulationScore¶
Plot multi-scale insulation scores from Juicer matrix.
plotMultiScaleInsulationScore [-h] [--num4norm NUM4NORM]
[--sizex SIZEX] [--sizey SIZEY]
matrix output resolution
Example:
plotInsulationScore WT/intrachromosomal/25000/observed.KR.chr7.matrix.gz MultiInsulationScore_WT.chr7.png 25000

Fig. 4.21 Multi-Insulation Score (chr7)¶
